| 九游集团品牌机器人 |
| CHUANGZE ROBOT |
外科手术的进步对急性和慢性疾病的管理,延长寿命和不断扩大生存范围都产生了重大影响。如图1所示,这些进步得益于诊断,成像和外科器械的持续技术发展。这些技术中,深度学习对推动术前手术规划尤其重要。手术规划中要根据现有的医疗记录来计划手术程序,而成像对于手术的成功至关重要。在现有的成像方式中,X射线,CT,超声和MRI是实际中最常用的方式。基于医学成像的常规任务包括解剖学分类,检测,分割和配准。
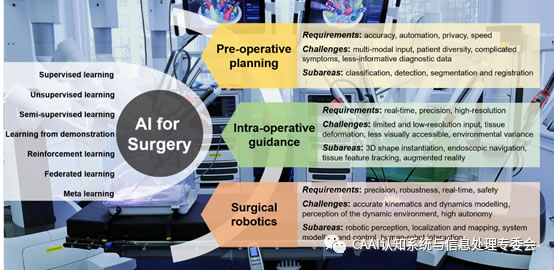
图1:概述了流行的AI技术,以及在术前规划,术中指导和外科手术机器人学中使用的AI的关键要求,挑战和子区域。
1、分类
分类输出输入的诊断值,该输入是单个或一组医学图像或器官或病变体图像。除了传统的机器学习和图像分析技术,基于深度学习的方法正在兴起[1]。对于后者,用于分类的网络架构由用于从输入层提取信息的卷积层和用于回归诊断值的完全连接层组成。
例如,有人提出了使用GoogleInception和ResNet架构的分类管道来细分肺癌,膀胱癌和乳腺癌的类型[2]。Chilamkurthy等证明深度学习可以识别颅内出血,颅骨骨折,中线移位和头部CT扫描的质量效应[3]。与标准的临床工具相比,可通过循环神经网络(RNN)实时预测心脏外科手术后患者的死亡率,肾衰竭和术后出血[4]。ResNet-50和Darknet-19已被用于对超声图像中的良性或恶性病变进行分类,显示出相似的灵敏度和更高的特异性[5]。
2、检测
检测通常以边界框或界标的形式提供感兴趣区域的空间定位,并且还可以包括图像或区域级别的分类。同样,基于深度学习的方法在检测各种异常或医学状况方面也显示出了希望。用于检测的DCNN通常由用于特征提取的卷积层和用于确定边界框属性的回归层组成。
为了从4D正电子发射断层扫描(PET)图像中检测前列腺癌,对深度堆叠的卷积自动编码器进行了训练,以提取统计和动力学生物学特征[6]。对于肺结节的检测,提出了具有旋转翻译组卷积(3D G-CNN)的3D CNN,具有良好的准确性,灵敏度和收敛速度[7]。对于乳腺病变的检测,基于深度Q网络扩展的深度强化学习(DRL)用于从动态对比增强MRI中学习搜索策略[8]。为了从CT扫描中检测出急性颅内出血并改善网络的可解释性,Lee等人[9]使用注意力图和迭代过程来模仿放射科医生的工作流程。
3、分割
分割可被视为像素级或体素级图像分类问题。由于早期作品中计算资源的限制,每个图像或卷积都被划分为小窗口,并且训练了CNN来预测窗口中心位置的目标标签。通过在密集采样的图像窗口上运行CNN分类器,可以实现图像或体素分割。例如,Deepmedic对MRI的多模式脑肿瘤分割显示出良好的性能[10]。但是,基于滑动窗口的方法效率低下,因为在许多窗口重叠的区域中会重复计算网络功能。由于这个原因,基于滑动窗口的方法最近被完全卷积网络(FCN)取代[11]。关键思想是用卷积层和上采样层替换分类网络中的全连接层,这大大提高了分割效率。对于医学图像分割,诸如U-Net [12][13]之类的编码器-解码器网络已显示出令人鼓舞的性能。编码器具有多个卷积和下采样层,可提取不同比例的图像特征。解码器具有卷积和上采样层,可恢复特征图的空间分辨率,并最终实现像素或体素密集分割。在[14]中可以找到有关训练U-Net进行医学图像分割的不同归一化方法的综述。
对于内窥镜胰管和胆道手术中的导航,Gibson等人 [15]使用膨胀的卷积和融合的图像特征在多个尺度上分割来自CT扫描的腹部器官。为了从MRI进行胎盘和胎儿大脑的交互式分割,将FCN与用户定义的边界框和涂鸦结合起来,其中FCN的最后几层根据用户输入进行了微调[16]。手术器械界标的分割和定位被建模为热图回归模型,并且使用FCN几乎实时地跟踪器械[17]。对于肺结节分割,Feng等通过使用候选筛选方法从弱标记的肺部CT中学习辨别区域来训练FCN,解决了需要精确的手动注释的问题[18]。Bai等提出了一种自我监督的学习策略,以有限的标记训练数据来提高U-Net的心脏分割精度[19]。
4、配准
配准是两个医学图像,体积或模态之间的空间对齐,这对于术前和术中规划都特别重要。传统算法通常迭代地计算参数转换,即弹性,流体或B样条曲线模型,以最小化两个医疗输入之间的给定度量(即均方误差,归一化互相关或互信息)。最近,深度回归模型已被用来代替传统的耗时和基于优化的注册算法。
示例性的基于深度学习的配准方法包括VoxelMorph,它通过利用基于CNN的结构和辅助分割来将输入图像对映射到变形场,从而最大化标准图像匹配目标函数[20]。提出了一个用于3D医学图像配准的端到端深度学习框架,该框架包括三个阶段:仿射变换预测,动量计算和非参数细化,以结合仿射配准和矢量动量参数化的固定速度场[21]。提出了一种用于多模式图像配准的弱监督框架,该框架对具有较高级别对应关系的图像(即解剖标签)进行训练,而不是用于预测位移场的体素级别转换[22]。每个马尔科夫决策过程都由经过扩张的FCN训练的代理商进行,以使3D体积与2D X射线图像对齐[23]。RegNet是通过考虑多尺度背景而提出的,并在人工生成的位移矢量场(DVF)上进行了培训,以实现非刚性配准[24]。3D图像配准也可以公式化为策略学习过程,其中将3D原始图像作为输入,将下一个最佳动作(即向上或向下)作为输出,并将CNN作为代理[25]。
参考文献:
[1] G. Litjens, T. Kooi, B. E.Bejnordi, A. A. A. Setio, F. Ciompi, M. Ghafoorian, J. A. Van Der Laak, B. VanGinneken, and C. I. Sa′nchez, “A survey on deep learning in medical image analysis,” Medical Image Analysis, vol. 42, pp. 60–88, 2017.
[2] P. Khosravi, E. Kazemi, M.Imielinski, O. Elemento, and I. Hajirasouliha, “Deep convolutional neural networks enable discrimination of heterogeneous digital pathology images,” EBioMedicine, vol. 27, pp. 317–328, 2018.
[3] S. Chilamkurthy, R. Ghosh, S.Tanamala, M. Biviji, N. G. Campeau, V. K. Venugopal, V. Mahajan, P. Rao, and P.Warier, “Deep learning algorithms for detection of critical findings in head CT scans: a retrospective study,” The Lancet, vol. 392, no. 10162, pp. 2388–2396,2018.
[4] A. Meyer, D. Zverinski, B.Pfahringer, J. Kempfert, T. Kuehne, S. H. Su¨ndermann, C. Stamm, T. Hofmann, V.Falk, and C. Eickhoff, “Machine learning for real-time prediction of complications in critical care: a retrospective study,” The Lancet RespiratoryMedicine, vol. 6, no. 12, pp. 905–914, 2018.
[5] X. Li, S. Zhang, Q. Zhang, X.Wei, Y. Pan, J. Zhao, X. Xin, C. Qin, X. Wang, J. Li et al., “Diagnosis of thyroid cancer using deep convolutional neural network models applied to sonographic images: a retrospective, multicohort, diagnostic study,” The LancetOncology, vol. 20, no. 2, pp. 193–201, 2019.
[6] E. Rubinstein, M. Salhov, M.Nidam-Leshem, V. White, S. Golan, J. Baniel, H. Bernstine, D. Groshar, and A.Averbuch, “Unsupervised tumor detection in dynamic PET/CT imaging of the prostate,” Medical Image Analysis, vol. 55, pp. 27–40, 2019.
[7] M. Winkels and T. S. Cohen,“Pulmonary nodule detection in CT scan with equivariant CNNs,” Medical image analysis, vol. 55, pp. 15–26, 2019.
[8] G. Maicas, G. Carneiro, A. P.Bradley, J. C. Nascimento, and I. Reid,“Deep reinforcement learning for active breast lesion detection from DCE-MRI,” in Proceedings of International Conference on Medical image computing and Computer-Assisted Intervention (MICCAI). Springer, 2017, pp.665–673.
[9] H. Lee, S. Yune, M. Mansouri,M. Kim, S. H. Tajmir, C. E. Guerrier, S. A. Ebert, S. R. Pomerantz, J. M.Romero, S. Kamalian et al., “An explainable deep-learning algorithm for the detection of acute intracranial hemorrhage from small datasets,” NatureBiomedical Engineering, vol. 3, no. 3, p. 173, 2019.
[10]K. Kamnitsas, C. Ledig, V. F.Newcombe, J. P. Simpson, A. D. Kane, D. K. Menon, D. Rueckert, and B. Glocker, “Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation,” Medical image analysis, vol. 36, pp. 61–78, 2017.
[11]J. Long, E. Shelhamer, and T.Darrell, “Fully convolutional networks for semantic segmentation,” in proceedings of the IEEE Conference on Computer Vision and Pattern Recognition,2015, pp. 3431–3440.
[12]O. Ronneberger, P. Fischer, and T. Brox, “U-Net: Convolutional networks for biomedical image segmentation,” in Proceedings of International Conference on Medical Image Computing and computer-Assisted Intervention (MICCAI). Springer, 2015, pp. 234–241.
[13]O. C¸i¸cek, A. Abdulkadir, S.S. Lienkamp, T. Brox, and O. Ronneberger,¨ “3D U-Net: learning dense volumetric segmentation from sparse annotation,” in Proceedings of InternationalConference on Medical Image Computing and Computer-Assisted Intervention(MICCAI). Springer, 2016, pp. 424–432.
[14]X.-Y. Zhou and G.-Z. Yang,“Normalization in training U-Net for 2D biomedical semantic segmentation,” IEEERobotics and Automation Letters, vol. 4, no. 2, pp. 1792–1799, 2019.
[15]E. Gibson, F. Giganti, Y. Hu,E. Bonmati, S. Bandula, K. Gurusamy, B. Davidson, S. P. Pereira, M. J.Clarkson, and D. C. Barratt, “Automatic multi-organ segmentation on abdominal CT with dense networks,” IEEE Transactions on Medical Imaging, vol. 37, no. 8,pp.1822–1834, 2018.
[16]G. Wang, W. Li, M. A. Zuluaga,R. Pratt, P. A. Patel, M. Aertsen, T. Doel, A. L. David, J. Deprest, S.Ourselin et al., “Interactive medical image segmentation using deep learning with image-specific fine-tuning,” IEEE Transactions on Medical Imaging, vol.37, no. 7, pp. 1562–1573, 2018.
[17]I. Laina, N. Rieke, C.Rupprecht, J. P. Vizca′ıno, A. Eslami, F. Tombari, and N. Navab, “Concurrentsegmentation and localization for tracking of surgical instruments,” in Proceedings of International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI).Springer, 2017, pp. 664–672.
[18]X. Feng, J. Yang, A. F. Laine,and E. D. Angelini, “Discriminative localization in CNNs for weakly-supervised segmentation of pulmonary nodules,” in Proceedings of International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI). Springer, 2017,pp. 568–576.
[19]W. Bai, C. Chen, G. Tarroni,J. Duan, F. Guitton, S. E. Petersen, Y. Guo, P. M. Matthews, and D. Rueckert,“Self-supervised learning for cardiac MR image segmentation by anatomical position prediction,” in International Conference on Medical Image Computing and ComputerAssisted Intervention. Springer, 2019, pp. 541–549.
[20]G. Balakrishnan, A. Zhao, M.R. Sabuncu, J. Guttag, and A. V. Dalca, “VoxelMorph: a learning framework for deformable medical image registration,” IEEE Transactions on Medical Imaging,2019.
[21]Z. Shen, X. Han, Z. Xu, and M.Niethammer, “Networks for joint affine and non-parametric image registration,”in Proceedings of the IEEE Conference on Computer Vision and pattern recognition (CVPR), 2019, pp. 4224–4233.
[22]Y. Hu, M. Modat, E. Gibson, W.Li, N. Ghavami, E. Bonmati, G. Wang, S. Bandula, C. M. Moore, M. Emberton etal., “Weaklysupervised convolutional neural networks for multimodal image registration,” Medical Image Analysis, vol. 49, pp. 1–13, 2018.
[23]S. Miao, S. Piat, P. Fischer,A. Tuysuzoglu, P. Mewes, T. Mansi, and R. Liao, “Dilated FCN for multi-agent2D/3D medical image registration,” in Proceedings of AAAI Conference on artificial intelligence, 2018.
[24]H. Sokooti, B. de Vos, F.Berendsen, B. P. Lelieveldt, I. Iˇsgum, and M. Staring, “Nonrigid image registration using multi-scale 3D convolutional neural networks,” in Proceedings of International Conference on Medical Image Computing and computer-Assisted Intervention (MICCAI). Springer, 2017, pp. 232–239.
[25]R. Liao, S. Miao, P. deTournemire, S. Grbic, A. Kamen, T. Mansi, and D. Comaniciu, “An artificial agent for robust image registration,” in Proceedings of AAAI Conference on Artificial Intelligence, 2017.
 |
| 机器人公司 机器人应用 智能医疗 物联网 机器人排名 机器人企业 教育机器人 机器人开发 独角兽 消毒机器人品牌 消毒机器人 |